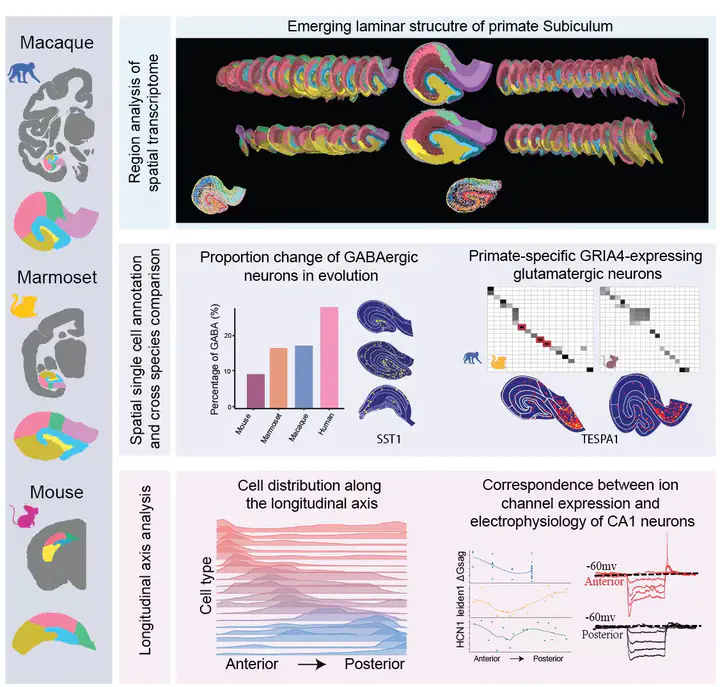
This project was conducted in collaboration with the Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, and aims to systematically characterize the cellular composition and spatial organization of hippocampal subregions across species. By integrating single-cell spatial transcriptomics and single-nucleus RNA sequencing data, high-resolution transcriptomic atlases of the macaque, marmoset, and mouse hippocampus were constructed, enabling cross-species comparisons of cell types and regional organization.
My contribution primarily focused on the integrative analysis of single-cell and spatial transcriptomic data, including cross-dataset cell-type label transfer to assign cellular identities to spatial data, optimization of spatial annotations, and visualization of the results.