Evolutionary convergence and divergence of hippocampal cytoarchitecture between rodents and primates revealed by single-cell spatial transcriptomics
Abstract
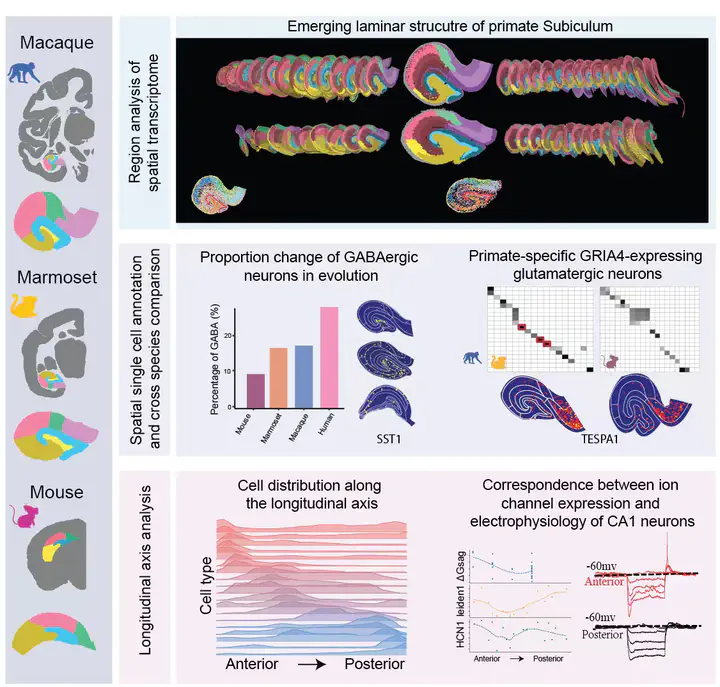
The hippocampus comprises subregions of distinct cell types critical for memory and cognition, but their gene expression profiles and spatial distribution patterns remain to be clarified. Using single-cell spatial transcriptomic analysis and single-nucleus RNA sequencing, we obtained transcriptome-based atlases for the macaque, marmoset, and mouse hippocampus. Cross-species comparison revealed primate- and lamina-specific glutamatergic cell types in the subicular complex, as well as enrichment of VIP-expressing GABAergic cells from mice to primates, including humans. Furthermore, we found reduced transcriptomic differences between CA3 and CA4 subregions and distinct longitudinal distributions of various cell types and expression of ion-channel genes, correlated with differences in electrophysiological properties of CA3, CA4, and CA1 neurons revealed by slice recording from marmosets and mice. Collectively, this cross-species study provides a molecular and cellular basis for understanding the evolution and function of the hippocampus.